C O da Costa-Luis
The following example may be launched interactively via any of the following:
Download and Preprocessing for PET-MR Simulations
This notebook will not re-download/re-process files if they already exist.
- Output data
~/.brainweb/subject_*.npz: dtype(shape):float32(127, 344, 344)
- Raw data
source
~/.brainweb/subject_*.bin.gz: dtype(shape):uint16(362, 434, 362)
- Install
pip install brainweb
from __future__ import print_function, division
%matplotlib notebook
import brainweb
from brainweb import volshow
import numpy as np
from os import path
from tqdm.auto import tqdm
import logging
logging.basicConfig(level=logging.INFO)
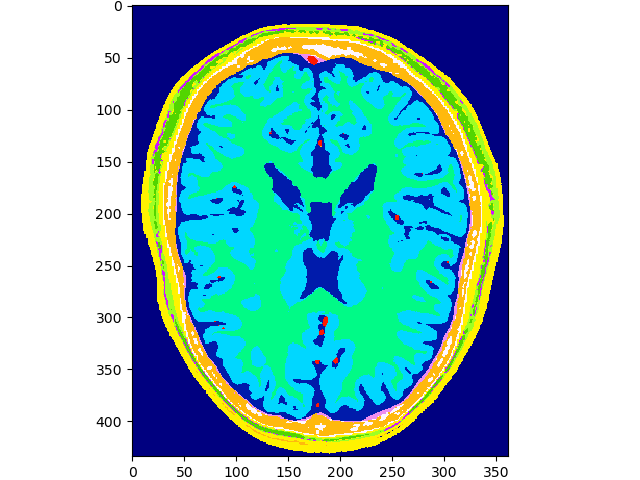
Raw Data
# download
files = brainweb.get_files()
# read last file
data = brainweb.load_file(files[-1])
# show last subject
print(files[-1])
volshow(data, cmaps=['gist_ncar']);
~/.brainweb/subject_54.bin.gz
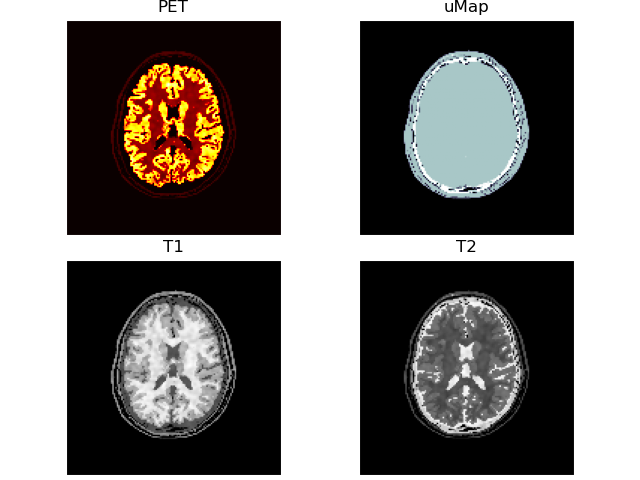
Transform
Convert raw image data:
- Siemens Biograph mMR resolution (~2mm) & dimensions (127, 344, 344)
- PET/T1/T2/uMap intensities
- randomised structure for PET/T1/T2
- $t (1 + g [2 G_\sigma(r) - 1])$, where
- $r = $
rand(127, 344, 344)$\in [0, 1)$, - Gaussian smoothing $\sigma = 1$,
- $g = 1$ for PET; 0.75 for MR, and
- $t$ = the PET or MR piecewise constant phantom
- $r = $
brainweb.seed(1337)
for f in tqdm(files, desc="mMR ground truths", unit="subject"):
vol = brainweb.get_mmr_fromfile(
f,
petNoise=1, t1Noise=0.75, t2Noise=0.75,
petSigma=1, t1Sigma=1, t2Sigma=1)
# show last subject
print(f)
volshow([vol['PET' ][:, 100:-100, 100:-100],
vol['uMap'][:, 100:-100, 100:-100],
vol['T1' ][:, 100:-100, 100:-100],
vol['T2' ][:, 100:-100, 100:-100]],
cmaps=['hot', 'bone', 'Greys_r', 'Greys_r'],
titles=["PET", "uMap", "T1", "T2"]);
~/.brainweb/subject_54.bin.gz
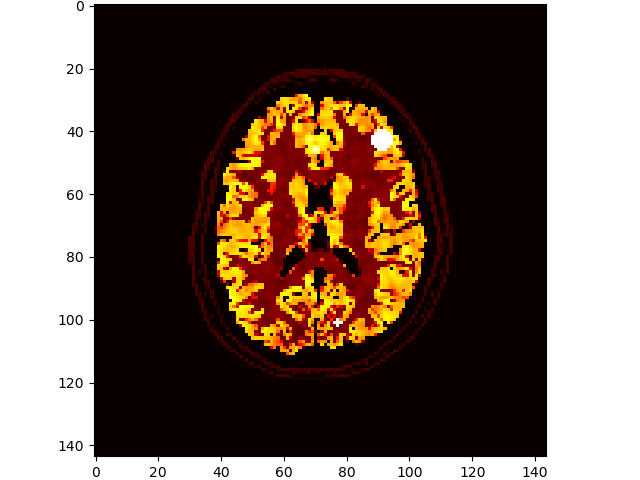
# add some lesions
brainweb.seed(1337)
im3d = brainweb.add_lesions(vol['PET'])
volshow(im3d[:, 100:-100, 100:-100], cmaps=['hot']);